OLFACTORYRECEPTORS Example of use of HMM in sequence analysis
Contents
Introduction
Proteins are composed of a sequence of amino acids. These aminoacids have various atomic compositions and structures that lead to different properties. The first part of this demo focuses on the property of hydrophobicity.
Olfactory receptors (OR) are part of a family of proteins that have 7 transmembrane regions. That is they pass through the cell membrane 7 times. The interior of the cell membrane is hydrophobic while both the exterior and interior of the cell are hydrophilic. Therefore, the regions of the protein that pass through the membrane should contain mostly hydrophobic amino acids while the portion outside of the membrane should be mostly hydrophilic.
Segmenting odorant receptors
You will create a segmentation Hidden Markov Model to predict the transmembrane (hydrophobic) regions of an olfactory receptor (OR) protein. In this example you will consider 347 OR due to Zozulya, et al., 2001. You can read the 347 sequence of amino acids with the MATLAB function fastaread
orseqs = fastaread('347OR.fasta');
Considering only the first sequence, you can plot various property of that using the tool MATLAB proteinplot.
or1 = orseqs(1).Sequence proteinplot(or1)
or1 = MPNSTTVMEFLLMRFSDVWTLQILHSASFFMLYLVTLMGNILIVTVTTCDSSLHMPMYFFLRNLSILDACYISVTVPTSCVNSLLDSTTISKAGCVAQVFLVVFFVYVELLFLTIMAHDRYVAVCQPLHYPVIVNSRICIQMTLASLLSGLVYAGMHTGSTFQLPFCRSNVIHQFFCDIPSLLKLSCSDTFSNEVMIVVSALGVGGGCFIFIIRSYIHIFSTVLGFPRGADRTKAFSTCIPHILVVSVFLSSCSSVYLRPPAIPAATQDLILSGFYSIMPPLFNPIIYSLRNKQIKVAIKKIMKRIFYSENV
In the protein plot window, you must change the selected property to hydrophobicity (Kyte & Doolittle). Under Edit -> Change Configuration, you must change the window size to 19.0. Now you need to export the figure so that you can plot the results of the model over it. To do this, select File -> Export Figure. Then hold the figure for future use. Here the final profile is loaded for simplicity.
open('hydrophobicity.fig'); % Comment this if you want to use the proteinplot hold on
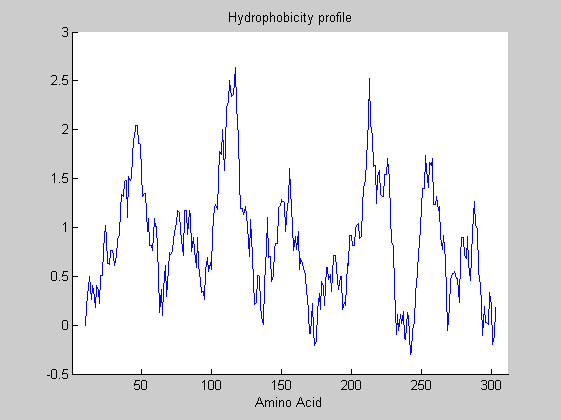
Although you can pick out a number of hydrophobic and hydrophilic peaks in this graph simply by eye, an HMM can help to delineate exactly the various regions of interest. A model is assumed that consider the protein sequence being generated by a stochastic process that alternates between two hidden states: “out of the membrane” and “in the membrane”. The HMM is trained using 20 OR sequences.
for i=1:20 intseqs(i) = {aa2int(orseqs(i).Sequence)}; end
The probability transition matrix is 2X2 for the 2 states: out of the membrane and in the membrane. You must enter an estimate of the transition probabilities as initial guess.
T = [0.95 0.05;
0.05 0.95];
The probability emission matrix is 2X20 (2 states and 20 amino acids). Also in this case you must enter the initial guesses of the emission probabilities. These guesses of emission are based on the hydrophobicity of the amino acids. For example, in the ‘in the membrane’ state, a hydrophilic amino acids has a higher probability of emission that one that is hydrophobic.
E = [0.018 0.067 0.067 0.067 0.018 0.067 0.067 0.067 0.067 0.01 0.01 0.067 0.018 0.018 0.067 0.067 0.067 0.067 0.067 0.01;
0.114 0.007 0.007 0.007 0.114 0.007 0.007 0.025 0.007 0.114 0.114 0.007 0.114 0.114 0.025 0.025 0.025 0.025 0.025 0.114];
Now, starting from this initial guesses the true emission and transition matrices must be estimated from our sequences using the EM algorithm. The Matlab function hmmtrain is used to this purpose.
[estT, estE] = hmmtrain(intseqs, T, E)
estT =
0.7338 0.2662
0.0711 0.9289
estE =
Columns 1 through 11
0.0820 0.0971 0.0657 0.0408 0.0000 0.0429 0.0553 0.0555 0.0187 0.0242 0.0715
0.0626 0.0181 0.0188 0.0248 0.0464 0.0153 0.0122 0.0416 0.0306 0.0902 0.1542
Columns 12 through 20
0.1480 0.0874 0.0000 0.0366 0.0818 0.0228 0.0045 0.0000 0.0653
0.0000 0.0424 0.0857 0.0427 0.0911 0.0763 0.0072 0.0583 0.0814
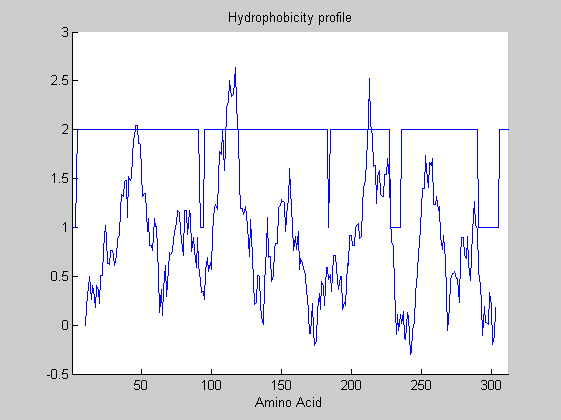
Finally the Viterbi algorithm can be used to detemine the states path, so to automatically segment the protein into its component regions. The MATLAB function hmmviterbi is used, receiving as input the emission and transition matrices previously estimated from hmmtrain. You can also plot the states over the hydrophobicity plot.
estimatedStates = hmmviterbi(aa2int(or1),estT,estE);
plot(estimatedStates)
hold off
Profile HMMs for odorant receptors
You will now turn to protein families and multiple alignment. Protein families are groups of proteins that have similar structure and function. Profile HMMs for specific families can be developed from the multiple alignment of members of the family. Profile HMMs create a useful position-based scoring system. Then, homologues can be compared back to the HMM. In MATLAB, you can use profile HMM to perform multiple sequence alignments.
Profile HMMs can be found for many protein families and the PFAM website.
web('http://www.sanger.ac.uk/Software/Pfam')
A first question to be answerd is to detemine which pHMM must be used for the olfactory receptors. You can use the first OR sequence and a randomized version of that to compare to the pHMMs.
randor1 = randseq(length(or1), 'fromstructure', aacount(or1));
There are over 7000 pHMMs available at the PFAM site. You can search all the HMMs by entering the sequence in the ‘search by protein sequence’ on the PFAM site. For sake of time, we will only compare the sequences to the first 4 pHMMs. The MATLAB function hmmprofalign aligns the sequences to the selected profile HMM while gethmmprof retrieves the model. For PFAM accession number PF00001, you must simply enter gethmmprof(1).
seqs = {or1,randor1};
for i = 1:4
for j = 1:2
[score(i,j)] = hmmprofalign(gethmmprof(i), seqs(j));
end
end
score
score = 111.2758 -112.1583 -178.5775 -160.0956 -157.0018 -161.5690 -97.2529 -101.9993
You can see that the fake protein did not show a good alignment with any of the HMMs. The real olfactory receptor matches PF00001. You will use this HMM to align the olfactory receptors.
Therefore, first of all, you should get the pHMM from the PFAM database. Then you can retrieve multiple aligned sequences from the PFAM database using the MATLAB function gethmmalignment.
hmm7tm = gethmmprof(1); seqs = gethmmalignment(1, 'type', 'seed'); disp([char(seqs.Header) char(seqs.Sequence)])
O10J1_HUMAN/52-300 GNIIIVTIIRIDLHLH....TPMYFFLSMLSTSETVYTLVILPRMLSSLVG........MSQPMSL...AGCATQMFFFVTFGITNCFLLTAMGYDRYVAICNPLRYMVIMN..KRLRIQLVLGACSIGLIVAITQVTS.VFRLPFC.ARK.......VPHFFCDIR...............PVMKLSCIDTTVNEILTLIISVLVLVVPMGLVFISYVLIIS...................................................................................................................................TILKIASVEGRKKAFATCASHLTVVIVHYSCASIAYLKPKSENT............REHDQLISVTYTVITPLLNPVVY OLF15_MOUSE/41-290 GNLTIILLSRLDARLH....TPMYFFLSNLSSLDLAFTTSSVPQMLKNLWG........PDKTISY...GGCVTQLYVFLWLGATECILLVVMAFDRYVAVCRPLHYMTVMN..PRLCWGLAAISWLGGLGNSVIQSTF.TLQLPFCGHRK.......VDNFLCEVP...............AMIKLACGDTSLNEAVLNGVCTFFTVVPVSVILVSYCFIAQ...................................................................................................................................AVMKIRSVEGRRKAFNTCVSHLVVVFLFYGSAIYGYLLPAKSSN............QSQGKFISLFYSVVTPMVNPLIY OL287_RAT/44-293 GNLAIISLVGAHRCLQ....TPMYFFLCNLSFLEIWFTTACVPKTLATFAP........RGGVISL...AGCATQMYFVFSLGCTEYFLLAVMAYDRYLAICLPLRYGGIMT..PGLAMRLALGSWLCGFSAITVPATL.IARLSFCGSRV.......INHFFCDIS...............PWIVLSCTDTQVVELVSFGIAFCVILGSCGITLVSYAYIIT...................................................................................................................................TIIKIPSARGRHRAFSTCSSHLTVVLIWYGSTIFLHVRTSVESS............LDLTKAITVLNTIVTPVLNPFIY OLF1_CHICK/41-290 TNLGLIALISVDLHLQ....TPMYIFLQNLSFTDAAYSTVITPKMLATFLE........ERKTISY...VGCILQYFSFVLLTVTESLLLAVMAYDRYVAICKPLLYPSIMT..KAVCWRLVESLYFLAFLNSLVHTSG.LLKLSFCYSNV.......VNHFFCDIS...............PLFQISSSSIAISELLVIISGSLFVMSSIIIILISYVFIIL...................................................................................................................................TVVMIRSKDGKYKAFSTCTSHLMAVSLFHGTVIFMYLRPVKLFS............LDTDKIASLFYTVVIPMLNPLIY GU27_RAT/22-271 GNLLIILAVSSNSHLH....NLMYFFLSNLSFVDICFISTTIPKMLVNIHS........QTKDISY...IECLSQVYFLTTFGGMDNFLLTLMACDRYVAICHPLNYTVIMN..LQLCALLILMFWLIMFCVSLIHVLLMNELNFSRG.............TEIPHFFCELA..........QVLKVANSDTHINNVFMYVVTSLLGLIPMTGILMSYSQIAS...................................................................................................................................SLLKMSSSVSKYKAFSTCGSHLCVVSLFYGSATIVYFCSSVLHS............THKKMIASLMYTVISPMLNPFIY MRGRF_RAT/61-291 GNGLVLWFFGFSIKRT.....PFSIYFLHLASADGIYLFSKAVIALLNMGT........FLGSFPD...YVRRVSRIVGLCTFFAGVSLLPAISIERCVSVIFPMWYWRRRP..KRLSAGVCALLWLLSFLVTSIHNYFCMFLGHEASG............TACLNMDISLG................ILLFFLFCPLMVLPCLALILHVECRARR................................................................................................................................................RQRSAKLNHVVLAIVSVFLVSSIYLGIDWFLFWVFQIP............APFPEYVTDLCICINSSAKPIVY OPSB_HUMAN/51-303 LNAMVLVATLRYKKLR....QPLNYILVNVSFGGFLLCIFSVFPVFVASCNG........YFVFGR...HVCALEGFLGTVAGLVTGWSLAFLAFERYIVICKPFGNFRFSS...KHALTVVLATWTIGIGVSIPPFFG.WSRFIPEG...........LQCSCGPDWYTVG....TKYRSESYTWFLFIFCFIVPLSLICFSYTQLLRALKAVAAQQQE........................................................................................................................................SATTQKAEREVSRMVVVMVGSFCVCYVPYAAFAMYMVNNRNHG..........LDLRLVTIPSFFSKSACIYNPIIY OPS3_DROME/75-338 GNGLVIWVFSAAKSLR....TPSNILVINLAFCDFMMMVKTPIFIYNSFHQG.........YALGH...LGCQIFGIIGSYTGIAAGATNAFIAYDRFNVITRPMEGKMTHG....KAIAMIIFIYMYATPWVVACYTETWGRFVPEG...........YLTSCTFDYLTDN......FDTRLFVACIFFFSFVCPTTMITYYYSQIVGHVFSHEKALRDQAKKMNV..........................................................................................................................ESLRSNVDKNKETAEIRIAKAAITICFLFFCSWTPYGVMSLIGAFGDKT..........LLTPGATMIPACACKMVACIDPFVY OPSD_LOLFO/51-315 GNGVVIYLFTKTKSLQ....TPANMFIINLAFSDFTFSLVNGFPLMTISCFM.......KYWVFGN...AACKVYGLIGGIFGLMSIMTMTMISIDRYNVIGRPMSASKKMS..HRKAFIMIIFVWIWSTIWAIGPIFGWGAYTLEGV............LCNCSFDYITRD......TTTRSNILCMYIFAFMCPIVVIFFCYFNIVMSVSNHEKEMAAMAKRLN............................................................................................................................AKELRKAQAGANAEMKLAKISIVIVTQFLLSWSPYAVVALLAQFGPIEW..........VTPYAAQLPVMFAKASAIHNPMIY OPS1_DROME/67-329 GNGVVIYIFATTKSLR....TPANLLVINLAISDFGIMITNTPMMGINLYF........ETWVLGP...MMCDIYAGLGSAFGCSSIWSMCMISLDRYQVIVKGMAGRPMTIP...LALGKIAYIWFMSSIWCLAPAFGWSRYVPEGN............LTSCGIDYLERDWN......PRSYLIFYSIFVYYIPLFLICYSYWFIIAAVSAHEKAMREQAKKMNV...........................................................................................................................KSLRSSEDAEKSAEGKLAKVALVTITLWFMAWTPYLVINCMGLFKFEG...........LTPLNTIWGACFAKSAACYNPIVY V2R_HUMAN/54-325 SNGLVLAALARRGRRGH..WAPIHVFIGHLCLADLAVALFQVLPQLAWKAT.........DRFRGP..DALCRAVKYLQMVGMYASSYMILAMTLDRHRAICRPMLAYRHGS..GAHWNRPVLVAWAFSLLLSLPQLFIFAQRNVEGGSG..........VTDCWACFAEPWG.......RRTYVTWIALMVFVAPTLGIAACQVLIFREIHASLVPGPSERPGGRRRG.......................................................................................................................RRTGSPGEGAHVSAAVAKTVRMTLVIVVVYVLCWAPFFLVQLWAAWDPEAP..........LEGAPFVLLMLLASLNSCTNPWIY FSHR_BOVIN/379-626 GNILVLVILITSQYKL....TVPRFLMCNLAFADLCIGIYLLLIASVDVHTKTEYHNYAIDWQTG....AGCDAAGFFTVFASELSVYTLTAITLERWHTITHAMQLECKVQ..LRHAASIMLVGWIFAFAVALFPIFGISSYMKVS...............ICLPMDIDSP........LSQLYVMSLLVLNVLAFVVICGCYTHIYLTVRNPNIT..............................................................................................................................................SSSSDTKIAKRMAMLIFTDFLCMAPISFFAISASLKVPL..........ITVSKSKILLVLFYPINSCANPFLY TRFR_HUMAN/42-320 GNIMVVLVVMRTKHMR....TPTNCYLVSLAVADLMVLVAAGLPNITDSIYG........SWVYGY...VGCLCITYLQYLGINASSCSITAFTIERYIAICHPIKAQFLCT..FSRAKKIIIFVWAFTSLYCMLWFFLLDLNISTYKD.........AIVISCGYKISRN........YYSPIYLMDFGVFYVVPMILATVLYGFIARILFLNPIPSDPKENSKTWKNDSTH..............................................................................................................QNTNLNVNTSNRCFNSTVSSRKQVTKMLAVVVILFALLWMPYRTLVVVNSFLSSP..........FQENWFLLFCRICIYLNSAINPVIY NTR1_HUMAN/80-364 GNTVTAFTLARKKSLQS.LQSTVHYHLGSLALSDLLTLLLAMPVELYNFIWV......HHPWAFGD...AGCRGYYFLRDACTYATALNVASLSVERYLAICHPFKAKTLMS..RSRTKKFISAIWLASALLAVPM.LFTMGEQNRSADGQH....AGGLVCTPTIHTATVK..........VVIQVNTFMSFIFPMVVISVLNTIIANKLTVMVRQAAEQGQVCTVGG......................................................................................................................EHSTFSMAIEPGRVQALRHGVRVLRAVVIAFVVCWLPYHVRRLMFCYISDE...QWTPFLYDFYHYFYMVTNALFYVSSTINPILY NPY1R_HUMAN/57-320 GNLALIIIILKQKEMR....NVTNILIVNLSFSDLLVAIMCLPFTFVYTLMD........HWVFGE...AMCKLNPFVQCVSITVSIFSLVLIAVERHQLIINPRGWRPNNR....HAYVGIAVIWVLAVASSLPFLIYQVMTDEPFQNVTLD...AYKDKYVCFDQFPSDS.......HRLSYTTLLLVLQYFGPLCFIFICYFKIYIRLKRRNNMMDKMR.....................................................................................................................................DNKYRSSETKRINIMLLSIVVAFAVCWLPLTIFNTVFDWNHQIIA.......TCNHNLLFLLCHLTAMISTCVNPIFY GPR83_MOUSE/88-345 GNVLVCHVIFKNQRMH....SATSLFIVNLAVADIMITLLNTPFTLVRFVN........STWVFGK...GMCHVSRFAQYCSLHVSALTLTAIAVDRHQVIMHPLKPRISIT....KGVIYIAVIWVMATFFSLPHAICQKLFTFKYSED........IVRSLCLPDFPEPAD.....LFWKYLDLATFILLYLLPLFIISVAYARVAKKLWLCNTIGDVTT....................................................................................................................................EQYLALRRKKKTTVKMLVLVVVLFALCWFPLNCYVLLLSSKAIH...........TNNALYFAFHWFAMSSTCYNPFIY NK1R_CAVPO/49-305 GNVVVMWIILAHKRMR....TVTNYFLVNLAFAEASMAAFNTVVNFTYAVHN........EWYYGL...FYCKFHNFFPIAAVFASIYSMTAVAFDRYMAIIHPLQPRLSAT....ATKVVICVIWVLALLLAFPQGYYSTTETMPGR.............VVCMIEWPSHP....DKIYEKVYHICVTVLIYFLPLLVIGYAYTVVGITLWASEIPGDSSD.....................................................................................................................................RYHEQVSAKRKVVKMMIVVVCTFAICWLPFHIFFLLPYINPDLY.......LKKFIQQVYLAIMWLAMSSTMYNPIIY TLR1_DROME/100-363 GNGIVLWIVTGHRSMR....TVTNYFLLNLSIADLLMSSLNCVFNFIFMLN........SDWPFGS...IYCTINNFVANVTVSTSVFTLVAISFDRYIAIVHPLKRRTSRR....KVRIILVLIWALSCVLSAPCLLYSSIMTKHYYNGKSR...TVCFMMWPDGRYPTSM.......ADYAYNLIILVLTYGIPMIVMLICYSLMGRVLWGSRSIGENTD.....................................................................................................................................RQMESMKSKRKVVRMFIAIVSIFAICWLPYHLFFIYAYHNNQV.......ASTKYVQHMYLGFYWLAMSNAMVNPLIY NPYR_DROME/122-383 GNGTVCYIVYSTPRMR....TVTNYFIASLAIGDILMSFFCVPSSFISLFIL.......NYWPFGL...ALCHFVNYSQAVSVLVSAYTLVAISIDRYIAIMWPLKPRITKR....YATFIIAGVWFIALATALPIPIVSGLDIPMSP..WH....TKCEKYICREMWPSRT.......QEYYYTLSLFALQFVVPLGVLIFTYARITIRVWAKRPPGEAET....................................................................................................................................NRDQRMARSKRKMVKMMLTVVIVFTCCWLPFNILQLLLNDEEFAHW........DPLPYVWFAFHWLAMSHCCYNPIIY CCKAR_HUMAN/58-370 GNTLVITVLIRNKRMR....TVTNIFLLSLAVSDLMLCLFCMPFNLIPNLLK........DFIFGS...AVCKTTTYFMGTSVSVSTFNLVAISLERYGAICKPLQSRVWQT..KSHALKVIAATWCLSFTIMTPYPIYSNLVPFTKNNNQ........TANMCRFLLPNDV.......MQQSWHTFLLLILFLIPGIVMMVAYGLISLELYQGIKFEASQKKSAKERKPSTTSSGKYEDSDGCYLQK.................................................................................TRPPRKLELRQLSTGSSSRANRIRSNSSAANLMAKKRVIRMLIVIVVLFFLCWMPIFSANAWRAYDTAS.......AERRLSGTPISFILLLSYTSSCVNPIIY BRS3_CAVPO/64-330 GNAILIKVFFKTKSMQ....TVPNIFITSLALGDLLLLLTCVPVDATHYLA........EGWLFGR...IGCKVLSFIRLTSVGVSVFTLTILSADRYKAVVKPLERQPSNA..ILKTCAKAGCIWIMSMIFALPEAIFSNVHTLRDPNKN.......MTSEWCAFYPVSEK......LLQEIHALLSFLVFYIIPLSIISVYYSLIARTLYKSTLNIPTEEQ...................................................................................................................................SHARKQVESRKRIAKTVLVLVALFALCWLPNHLLNLYHSFTHKAYE.....DSSAIHFIVTIFSRVLAFSNSCVNPFAL MC3R_MOUSE/55-299 ENILVILAVVRNGNLH....SPMYFFLCSLAAADMLVSLSNSLETIMIAVINSD..SLTLEDQFIQ...HMDNIFDSMICISLVASICNLLAIAIDRYVTIFYALRYHSIMT..VRKALTLIGVIWVCCGICGVMFIIYSESKM................VIVCLITMFFAM...........VLLMGTLYIHMFLFARLHVQRIAVLPPAGVVAPQ...............................................................................................................................................QHSCMKGAVTITILLGVFIFCWAPFFLHLVLIITCPTN.......PYCICYTAHFNTYLVLIMCNSVIDPLIY ACM1_HUMAN/42-418 GNLLVLISFKVNTELK....TVNNYFLLSLACADLIIGTFSMNLYTTYLLMG........HWALGT...LACDLWLALDYVASNASVMNLLLISFDRYFSVTRPLSYRAKRT..PRRAALMIGLAWLVSFVLWAPA.ILFWQYLVGERTVL.........AGQCYIQFLSQP..........IITFGTAMAAFYLPVTVMCTLYWRIYRETENRARELAALQGSETPGKGGGSSSSSERSQPGAEGSPETPPGRCCRCCRAPRLLQAYSWKEEEEEDEGSMESL........TSSEGEEPGSEVVIKMPMVDPEAQAPTKQPPRSSPNTVKRPTKKGRDRAGKGQKPRGKEQLAKRKTFSLVKEKKAARTLSAILLAFILTWTPYNIMVLVSTFCKDC...........VPETLWELGYWLCYVNSTINPMCY 5HT2A_CRIGR/91-380 GNILVIMAVSLEKKLQ....NATNYFLMSLAIADMLLGFLVMPVSMLTILYG.......YRWPLPS...KLCAVWIYLDVLFSTASIMHLCAISLDRYVAIQNPIHHSRFNS..RTKAFLKIIAVWTISVGVSMPIPVFGLQDDSKVFK...........QGSCLLADDNF.............VLIGSFVAFFIPLTIMVITYFLTIKSLQKEATLCVSDLSTRAKLASFSFLPQSSLS................................................................................................SEKLFQRSIHREPGSYTGRRTMQSISNEQKACKVLGIVFFLFVVMWCPFFITNIMAVICKESCN.......EHVIGALLNVFVWIGYLSSAVNPLVY 5HT5A_MOUSE/57-338 WNLLVLATILKVRTFH....RVPHNLVASMAISDVLVAVLVMPLSLVHELSG.......RRWQLGR...RLCQLWIACDVLCCTASIWNVTAIALDRYWSITRHLEYTLRTR..KRVSNVMILLTWALSTVISLAPLLFGWGETYSEP............SEECQVSREPS............YTVFSTVGAFYLPLWLVLFVYWKIYRAAKFRMGSRKTNSVSPVPEAVEVKNATQH....................................................................................................PQMVFTARHATVTFQTEGDTWREQKEQRAALMVGILIGVFVLCWFPFFVTELISPLCSWD...........VPAIWKSIFLWLGYSNSFFNPLIY HRH2_CANFA/35-288 GNVVVCLAVGLNRRLR....SLTNCFIVSLAITDLLLGLLVLPFSAFYQLS........CRWSFGK...VFCNIYTSLDVMLCTASILNLFMISLDRYCAVTDPLRYPVLIT..PVRVAVSLVLIWVISITLSFLSIHLGWNSRNETSSF..........NHTIPKCKVQVN.........LVYGLVDGLVTFYLPLLVMCITYYRIFKIARDQAKRIHHMG.....................................................................................................................................SWKAATIGEHKATVTLAAVMGAFIICWFPYFTVFVYRGLKGDD..........AINEAFEAVVLWLGYANSALNPILY DRD1_HUMAN/40-331 GNTLVCAAVIRFRHLR...SKVTNFFVISLAVSDLLVAVLVMPWKAVAEIAG........FWPFG....SFCNIWVAFDIMCSTASILNLCVISVDRYWAISSPFRYERKMT..PKAAFILISVAWTLSVLISFIPVQLSWHKAKPTSPS..........DGNATSLAETIDNC..DSSLSRTYAISSSVISFYIPVAIMIVTYTRIYRIAQKQIRRIAALERAAVHAKNCQTTT...........................................................................................................GNGKPVECSQPESSFKMSFKRETKVLKTLSVIMGVFVCCWLPFFILNCILPFCGSG.....ETQPFCIDSNTFDVFVWFGWANSSLNPIIY ADRB1_HUMAN/75-377 GNVLVIVAIAKTPRLQ....TLTNLFIMSLASADLVMGLLVVPFGATIVVW........GRWEYGS...FFCELWTSVDVLCVTASIETLCVIALDRYLAITSPFRYQSLLT..RARARGLVCTVWAISALVSFLPILMHWWRAESDE............ARRCYNDPKCCD....FVTN.RAYAIASSVVSFYVPLCIMAFVYLRVFREAQKQVKKIDSCERRFLGGPARPPSPSPSPVPAPAPP.....................................................................................PGPPRPAAAAATAPLANGRAGKRRPSRLVALREQKALKTLGIIMGVFTLCWLPFFLANVVKAFHREL...........VPDRLFVFFNWLGYANSAFNPIIY 5HT1R_DROME/179-507GNVLVCIAVCMVRKLR....RPCNYLLVSLALSDLCVALLVMPMALLYEVL........EKWNFGP...LLCDIWVSFDVLCCTASILNLCAISVDRYLAITKPLEYGVKRT..PRRMMLCVGIVWLAAACISLPP.LLILGNEHEDEEG..........QPICTVCQNFA............YQIYATLGSFYIPLSVMLFVYYQIFRAARRIVLEEKRAQTHLQQALNGTGSPSAPQAPPLGHTELASSGNGQRHSSVGN.....................................................TSLTYSTCGGLSSGGGALAGHGSGGGVSGSTGLLGSPHHKKLRFQLAKEKKASTTLGIIMSAFTVCWLPFFILALIRPFETMHV...........PASLSSLFLWLGYANSLLNPIIY 5HT7R_HUMAN/98-384 GNCLVVISVCFVKKLR....QPSNYLIVSLALADLSVAVAVMPFVSVTDLIG.......GKWIFGH...FFCNVFIAMDVMCCTASIMTLCVISIDRYLGITRPLTYPVRQN..GKCMAKMILSVWLLSASITLPP.LFGWAQNVNDDK.............VCLISQDFG............YTIYSTAVAFYIPMSVMLFMYYQIYKAARKSAAKHKFPGFPRVEPDSVIALNGIVKL.................................................................................................QKEVEECANLSRLLKHERKNISIFKREQKAATTLGIIVGAFTVCWLPFFLLSTARPFICGTSCSCI.......PLWVERTFLWLGYANSLINPFIY 5HT1B_HUMAN/66-369 SNAFVIATVYRTRKLH....TPANYLIASLAVTDLLVSILVMPISTMYTVT........GRWTLGQ...VVCDFWLSSDITCCTASILHLCVIALDRYWAITDAVEYSAKRT..PKRAAVMIALVWVFSISISLPP..FFWRQAKAEEEV...........SECVVNTDHIL...........YTVYSTVGAFYFPTLLLIALYGRIYVEARSRILKQTPNRTGKRLTRAQLITDSPGSTSSVTSINSR...............................................................................VPDVPSESGSPVYVNQVKVRVSDALLEKKKLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPICKDACWF.........HLAIFDFFTWLGYLNSLINPIIY 5HT1A_HUMAN/53-400 GNACVVAAIALERSLQ....NVANYLIGSLAVTDLMVSVLVLPMAALYQVL........NKWTLGQ...VTCDLFIALDVLCCTSSILHLCAIALDRYWAITDPIDYVNKRT..PRRAAALISLTWLIGFLISIPP.MLGWRTPEDRSDP...........DACTISKDHG............YTIYSTFGAFYIPLLLMLVLYGRIFRAARFRIRKTVKKVEKTGADTRHGASPAPQPKKSVNGESGSRNWRLGVESKAGGALCANGAVR...................................QGDDGAALEVIEVHRVGNSKEHLPLPSEAGPTPCAPASFERKNERNAEAKRKMALARERKTVKTLGIIMGTFILCWLPFFIVALVLPFCESSCHM.........PTLLGAIINWLGYSNSLLNPVIY DRD2_BOVIN/51-427 GNVLVCMAVSREKALQ....TTTNYLIVSLAVADLLVATLVMPWVVYLEVVG........EWKFSR...IHCDIFVTLDVMMCTASILNLCAISIDRYTAVAMPMLYNTRYSS.KRRVTVMIAIVWVLSFTISCPMLFG.LNNTDQNE...............CIIANPAF.............VVYSSIVSFYVPFIVTLLVYIKIYIVLRRRRKRVNTKRSSRAFRANLKAPLKGNCTHPEDMKLCTVIMKSNGSFPVNRRRVEAARRAQELEMEMLSSTSPPERTRYSPIPPSHHQLTLPDPSHHGLHSTPDSPAKPEKNGHAKTVNPKIAKIFEIQSMPNGKTRTSLKTMSRRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCN...........IPPVLYSAFTWLGYVNSAVNPIIY ADA1D_HUMAN/113-402GNLLVILSVACNRHLQ....TVTNYFIVNLAVADLLLSATVLPFSATMEVLG........FWAFGR...AFCDVWAAVDVLCCTASILSLCTISVDRYVGVRHSLKYPAIMT..ERKAAAILALLWVVALVVSVGP.LLGWKEPVPPD............ERFCGITEEAG............YAVFSSVCSFYLPMAVIVVMYCRVYVVARSTTRSLEAGVKRERGKASEVVLRIHCRGAAT...........................................................................................GADGAHGMRSAKGHTFRSSLSVRLLKFSREKKAAKTLAIVVGVFVLCWFPFFFVLPLGSLFPQLK..........PSEGVFKVIFWLGYFNSCVNPLIY 5HT6R_RAT/43-320 ANSLLIVLICTQPALR....NTSNFFLVSLFTSDLMVGLVVMPPAMLNALYG........RWVLAR...GLCLLWTAFDVMCCSASILNLCLISLDRYLLILSPLRYKLRMT..APRALALILGAWSLAALASFLPLLLGWHELGKARTPA.........PGQCRLLASLP............FVLVASGVTFFLPSGAICFTYCRILLAARKQAVQVASLTTGTAGQALETLQVP.........................................................................................................RTPRPGMESADSRRLATKHSRKALKASLTLGILLGMFFVTWLPFFVANIAQAVCDCIS............PGLFDVLTWLGYCNSTMNPIIY AA1R_BOVIN/26-288 GNVLVIWAVKVNQALR....DATFCFIVSLAVADVAVGALVIPLAILINIG..........PRTYF...HTCLKVACPVLILTQSSILALLAMAVDRYLRVKIPLRYKTVVT..PRRAVVAITGCWILSFVVGLTP.MFGWNNLSAVERDWLANGSVGEPVIECQFEKVIS.........MEYMVYFNFFVWVLPPLLLMVLIYMEVFYLIRKQLSKKVSASS...................................................................................................................................GDPQKYYGKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCH..........MPRILIYIAIFLSHGNSAMNPIVY PTAFR_CAVPO/32-293 ANGYVLWVFARLYPSKK..LNEIKIFMVNLTVADLLFLITLPLWIVYYSNQ........GNWFLPK...FLCNLAGCLFFINTYCSVAFLGVITYNRFQAVKYPIKTAQATT..RKRGIALSLVIWVAIVAAASYFLVMDSTNVVSNKAGS.......GNITRCFEHYEKGS.......KPVLIIHICIVLGFFIVFLLILFCNLVIIHTLLRQPVKQQ...........................................................................................................................................RNAEVRRRALWMVCTVLAVFVICFVPHHMVQLPWTLAELG...MWPSSNHQAINDAHQVTLCLLSTNCVLDPVIY PAR1_CRILO/122-374 LNILAIAVFVLKMKVK....KPAVVYMLHLAMADVLFVSVLPLKISYYFSG........SDWQFGS...GMCRFATAAFYCNMYASIMLMTVISIDRFLAVVYPIQSLSWRT..LGRANFTCLVIWVMAIMGVVPLLLKEQTTRVPGLN...........ITTCHDVLNETLLQG....FYSYYFSAFSAVFFLVPLIISTICYMSIIRCLSSSSVA..............................................................................................................................................NRSKKSRALFLSAAVFCVFIVCFGPTNVLLIMHYLLLSD......SPATEKAYFAYLLCVCVSSVSCCIDPLIY P2RY5_CHICK/31-288 ANCVAIYIFTFTLKVR....NETTTYMLNLAISDLLFVFTLPFRIYYFVVR.........NWPFGD...VLCKISVTLFYTNMYGSILFLTCISVDRFLAIVHPFRSKTLRT..KRNARIVCVAVWITVLAGSTPASFFQSTNRQNNTE...........QRTCFENFPEST....WKTYLSRIVIFIEIVGFFIPLILNVTCSTMVLRTLNKPLTLS............................................................................................................................................RNKLSKKKVLKMIFVHLVIFCFCFVPYNITLILYSLMRTQTWIN..CSVVTAVRTMYPVTLCIAVSNCCFDPIVY EBI2_HUMAN/48-308 GNLLALVVIVQ.NRKK...INSTTLYSTNLVISDILFTTALPTRIAYYAMG........FDWRIGD...ALCRITALVFYINTYAGVNFMTCLSIDRFIAVVHPLRYNKIKR..IEHAKGVCIFVWILVFAQTLPLLINPMSKQEAERI...........TCMEYPNFEETKS.......LPWILLGACFIGYVLPLIIILICYSQICCKLFRTAKQNPL.........................................................................................................................................TEKSGVNKKALNTIILIIVVFVLCFTPYHVAIIQHMIKKLRFSNFLECSQRHSFQISLHFTVCLMNFNCCMDPFIY US28_HCMVT/50-291 GNFLVIFTITWRRRIQ....CSGDVYFINLAAADLLFVCTLPLWMQYLL..........DHNSLAS...VPCTLLTACFYVAMFASLCFITEIALDRYYAIV....YMRYRP..VKQACLFSIFWWIFAVIIAIPHFMVVTKKDNQ.................CMTDYDYLE....VS.YPIILNVELMLGAFVIPLSVISYCYYRISRIVAVSQS.................................................................................................................................................RHKGRIVRVLIAVVLVFIIFWLPYHLTLFVDTLKLLKWI.SSSCEFERSLKRALILTESLAFCHCCLNPLLY CX3C1_RAT/49-294 GNLLVVLALTNSRKSK....SITDIYLLNLALSDLLFVATLPFWTHYLIS.........HEGLH.N...AMCKLTTAFFFIGFFGGIFFITVISIDRYLAIVLAANSMNNRT..VQHGVTISLGVWAAAILVASPQFMFTKRK.................DNECLGDYPEVL....QEIWPVLRNSEVNILGFVLPLLIMSFCYFRIVRTLFSCKN.................................................................................................................................................RKKARAIRLILLVVVVFFLFWTPYNIVIFLETLKFYNFF..PSCGMKRDLRWALSVTETVAFSHCCLNPFIY CCR1_HUMAN/51-301 GNILVVLVLVQYKRLK....NMTSIYLLNLAISDLLFLFTLPFWIDYKLK.........DDWVFGD...AMCKILSGFYYTGLYSEIFFIILLTIDRYLAIVHAVFALRART..VTFGVITSIIIWALAILASMPGLYF.SKTQWEFT............HHTCSLHFPHES....LREWKLFQALKLNLFGLVLPLLVMIICYTGIIKILLRRPN.................................................................................................................................................EKKSKAVRLIFVIMIIFFLFWTPYNLTILISVFQDFLFT..HECEQSRHLDLAVQVTEVIAYTHCCVNPVIY CCRL1_BOVIN/58-303 GNSTVVAIYAYYKKRR....TKTDVYILNLAVADLFLLFTLPFWAVNAVHG..........WVLGK...IMCKVTSALYTVNFVSGMQFLACISTDRYWAVTKAPSQSGVGK....PCWVICFCVWVAAILLSIPQLVFYTVNHKARCVPI.........FPYHLGTSMKAS...........IQILEICIGFIIPFLIMAVCYFITAKTLIKMPN.................................................................................................................................................IKKSQPLKVLFTVVIVFIVTQLPYNIVKFCQAIDIIYSL.ITDCDMSKRMDVAIQITESIALFHSCLNPVLY CCR7_HUMAN/75-326 GNGLVVLTYIYFKRLK....TMTDTYLLNLAVADILFLLTLPFWAYSAAKS..........WVFGV...HFCKLIFAIYKMSFFSGMLLLLCISIDRYVAIVQAVSAHRHRARVLLISKLSCVGIWILATVLSIPELLYSDLQRSSSEQ...........AMRCSLITEHVEA.......FITIQVAQMVIGFLVPLLAMSFCYLVIIRTLLQARN.................................................................................................................................................FERNKAIKVIIAVVVVFIVFQLPYNGVVLAQTVANFNIT.SSTCELSKQLNIAYDVTYSLACVRCCVNPFLY CXCR4_BOVIN/56-303 GNGLVILVMGYQKKLR....SMTDKYRLHLSVADLLFVLTLPFWAVDAVAN..........WYFGK...FLCKAVHVIYTVNLYSSVLILAFISLDRYLAIVHATNSQKPRK..LLAEKVVYVGVWLPAVLLTIPDLIFADIKEVDER.............YICDRFYPSDL.......WLVVFQFQHIVVGLLLPGIVILSCYCIIISKLSHSKG.................................................................................................................................................YQKRKALKTTVILILTFFACWLPYYIGISIDSFILLEII.QQGCEFESTVHKWISITEALAFFHCCLNPILY CXCR1_HUMAN/56-305 GNSLVMLVILYSRVGR....SVTDVYLLNLALADLLFALTLPIWAASKVNG..........WIFGT...FLCKVVSLLKEVNFYSGILLLACISVDRYLAIVHATRTLTQKR...HLVKFVCLGCWGLSMNLSLPFFLFRQAYHPNNSSP............VCYEVLGNDT.....AKWRMVLRILPHTFGFIVPLFVMLFCYGFTLRTLFKAHM.................................................................................................................................................GQKHRAMRVIFAVVLIFLLCWLPYNLVLLADTLMRTQVI.QETCERRNNIGRALDATEILGFLHSCLNPIIY CXCR5_HUMAN/68-322 GNVLVLVILERHRQTR....SSTETFLFHLAVADLLLVFILPFAVAEGSVG..........WVLGT...FLCKTVIALHKVNFYCSSLLLACIAVDRYLAIVHAVHAYRHRR..LLSIHITCGTIWLVGFLLALPEILFAKVSQGHHNNS..........LPRCTFSQENQA....ETHAWFTSRFLYHVAGFLLPMLVMGWCYVGVVHRLRQAQR................................................................................................................................................RPQRQKAVRVAILVTSIFFLCWSPYHIVIFLDTLARLKAV.DNTCKLNGSLPVAITMCEFLGLAHCCLNPMLY APJ_HUMAN/45-309 GNGLVLWTVFRSSREK...RRSADIFIASLAVADLTFVVTLPLWATYTYRD........YDWPFGT...FFCKLSSYLIFVNMYASVFCLTGLSFDRYLAIVRPVANARLRL..RVSGAVATAVLWVLAALLAMPVMVLRTTGDLENTT...........KVQCYMDYSMVATVSSEWAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAGHFRKER..........................................................................................................................................IEGLRKRRRLLSIIVVLVVTFALCWMPYHLVKTLYMLGSLLH...WPCDFDLFLMNIFPYCTCISYVNSCLNPFLY BKRB2_HUMAN/74-332 ENIFVLSVFCLHKSSC....TVAEIYLGNLAAADLILACGLPFWAITISNN........FDWLFGE...TLCRVVNAIISMNLYSSICFLMLVSIDRYLALVKTMSMGRMRG..VRWAKLYSLVIWGCTLLLSSPMLVFRTMKEYSDEGHN.........VTACVISYPSLI.......WEVFTNMLLNVVGFLLPLSVITFCTMQIMQVLRNNEMQKF...........................................................................................................................................KEIQTERRATVLVLVVLLLFIICWLPFQISTFLDTLHRLGI..LSSCQDERIIDVITQIASFMAYSNSCLNPLVY AGTR1_BOVIN/45-302 GNSLVVIVIYFYMKLK....TVASVFLLNLALADLCFLLTLPLWAVYTAMEY........RWPFGN...YLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRT..MLVAKVTCIIIWLLAGLASLPTIIHRNVFFIENTN...........ITVCAFHYESQN.....STLPVGLGLTKNILGFLFPFLIILTSYTLIWKTLKKAYEIQ............................................................................................................................................KNKPRKDDIFKIILAIVLFFFFSWVPHQIFTFMDVLIQLGL..IRDCKIEDIVDTAMPITICLAYFNNCLNPLFY AGTR2_MOUSE/61-318 VNIVVVSLFCCQKGPK....KVSSIYIFNLALADLLLLATLPLWATYYSYR........YDWLFGP...VMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFLSQRRNP...WQASYVVPLVWCMACLSSLPTFYFRDVRTIEYLG...........VNACIMAFPPEK....YAQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNSYG............................................................................................................................................KNRITRDQVLKMAAAVVLAFIICWLPFHVLTFLDALTWMGI..INSCEVIAVIDLALPFAILLGFTNSCVNPFLY C5AR_CANFA/55-302 GNFLVVWVTGFEVRRT.....INAIWFLNLAVADLLSCLALPILFSSIVQQG........YWPFGN...AACRILPSLILLNMYASILLLTTISADRFVLVFNPIWCQNYRG..PQLAWAACSVAWAVALLLTVPSFIFRGVHTEYFPF...........WMTCGVDYSGVG.....VLVERGVAILRLLMGFLGPLVILSICYTFLLIRTWSRKA.................................................................................................................................................TRSTKTLKVVVAVVVSFFVLWLPYQVTGMMMALFYKHS......ESFRRVSRLDSLCVAVAYINCCINPIIY SSR1_HUMAN/75-323 GNSMVIYVILRYAKMK....TATNIYILNLAIADELLMLSVPFLVTSTLL.........RHWPFGA...LLCRLVLSVDAVNMFTSIYCLTVLSVDRYVAVVHPIKAARYRR..PTVAKVVNLGVWVLSLLVILPIVVFSRTAANSDG............TVACNMLMPEPA.....QRWLVGFVLYTFLMGFLLPVGAICLCYVLIIAKMRMVALKAGW.........................................................................................................................................QQRKRSERKITLMVMMVVMVFVICWMPFYVVQLVNVFAEQD....D........ATVSQLSVILGYANSCANPILY OPRD_MOUSE/66-318 GNVLVMFGIVRYTKLK....TATNIYIFNLALADALATSTLPFQSAKYLM.........ETWPFGE...LLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRT..PAKAKLINICIWVLASGVGVPIMVM.AVTQPRDGAVV.......CMLQFPSPS..........WYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS.........................................................................................................................................KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDINRRDPL.......VVAALHLCIALGYANSSLNPVLY RDC1_CANFA/61-315 ANSVVVWVNIQAKTTG....YDTHCYILNLAIADLWVVVTIPVWVVSLVQHN........QWPMGE...LTCKITHLIFSINLFGSIFFLTCMSVDRYLSITYFASTSSRRK..KVVRRAVCVLVWLLAFCVSLPDTYYLKTVTSASNN...........ETYCRSFYPEHS....VKEWLISMELVSVVLGFAIPFCVIAVFYCLLARAISASSD.................................................................................................................................................QEKQSSRKIIFSYVVVFLVCWLPYHVVVLLDIFSILHYI.PFTCQLENFLFTALHVTQCLSLVHCCVNPVLY ADMR_RAT/66-316 ENVLVICVNCR.RSGR...VGMLNLYILNMAVADLGIILSLPVWMLEVMLE........YTWLWGS...FSCRFIHYFYLANMYSSIFFLTCLSIDRYVTLTNTSPSWQRHQ..HRIRRAVCAGVWVLSAIIPLPEVVHIQLLDGSEP..............MCLFLAPFET....YSAWALAVALSATILGFLLPFPLIAVFNILSACRLRRQGQ.................................................................................................................................................TESRRHCLLMWAYIVVFVICWLPYHVTMLLLTLHTTHI..FLHCNLVNFLYFFYEIIDCFSMLHCVANPILY US27_HCMVA/47-294 LNVLVITTILYYRRKK...KSPSDTYICNLAVADLLIVVGLPFFLEYAKH.........HPKLSRE...VVCSGLNACFYICLFAGVCFLINLSMDRYCVIVWGVELNRVRN..NKRATCWVVIFWILAVLMGMPHYLMYSHTNNECVGEF.........ANETSGWFPVF............LNTKVNICGYLAPIALMAYTYNRMVRFIINYVG.................................................................................................................................................KWHMQTLHVLLVVVVSFASFWFPFNLALFLESIRLLAGV..YNDTLQNVIIFCLYVGQFLAYVRACLNPGIY EDG1_HUMAN/62-310 ENIFVLLTIWKTKKFH....RPMYYFIGNLALSDLLAGVAYTANLLLSGAT.........TYKLTP...AQWFLREGSMFVALSASVFSLLAIAIERYITMLKMKLHNGSNN...FRLFLLISACWVISLILGGLPIMGWNCISALSS...............CSTVLPLYH..........KHYILFCTTVFTLLLLSIVILYCRIYSLVRTRSRRLTFRKN...................................................................................................................................ISKASRSSENVALLKTVIIVLSVFIACWAPLFILLLLDVGCKVKT.........CDILFRAEYFLVLAVLNSGTNPIIY CNR2_HUMAN/50-299 ENVAVLYLILSSHQLR...RKPSYLFIGSLAGADFLASVVFACSFVNFHVF.........HGVDSK...AVFLLKIGSVTMTFTASVGSLLLTAIDRYLCLRYPPSYKALLT..RGRALVTLGIMWVLSALVSYLPLMGWTCCPRP.................CSELFPLIP..........NDYLLSWLLFIAFLFSGIIYTYGHVLWKAHQHVASLSGHQDR.................................................................................................................................QVPGMARMRLDVRLAKTLGLVLAVLLICWFPVLALMAHSLATTLS.....DQ.....VKKAFAFCSMLCLINSMVNPVIY CNR1_HUMAN/133-397 ENLLVLCVILHSRSLR...CRPSYHFIGSLAVADLLGSVIFVYSFIDFHVF.........HRKDSR...NVFLFKLGGVTASFTASVGSLFLTAIDRYISIHRPLAYKRIVT..RPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSV...............CSDIFPHID..........ETYLMFWIGVTSVLLLFIVYAYMYILWKAHSHAVRMIQRGTQKSIIIH....................................................................................................................TSEDGKVQVTRPDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMN.....KL.....IKTVFAFCSMLCLLNSTVNPIIY UL33_HCMVA/48-306 LNAIVLITQLLTNRVLG..YSTPTIYMTNLYSTNFLTLTVLPFIVLSNQWLL..........PAGV...ASCKFLSVIYYSSCTVGFATVALIAADRYRVLH..KRTYARQS..YRSTYMILLLTWLAGLIFSVPAAVYTTVVMHHDANDTN....NTNGHATCVLYFVAEE....VHTVLLSWKVLLTMVWGAAPVIMMTWFYAFFYSTVQRTSQ.................................................................................................................................................KQRSRTLTFVSVLLISFVALQTPYVSLMIFNSYATTAW..PMQCEHLTLRRTIGTLARVVPHLHCLINPILY TA2R_HUMAN/41-308 SNLLALSVLAGARQGGSHTRSSFLTFLCGLVLTDFLGLLVTGTIVVSQHAAL.......FEWHAVDPGCRLCRFMGVVMIFFGLSPLLLGAAMASERYLGITRPFSRPAVAS..QRRAWATVGLVWAAALALGLLPLLGVGRYTVQYP............GSWCFLTLGAES......GDVAFGLLFSMLGGLSVGLSFLLNTVSVATLCHVYHGQEAAQ.........................................................................................................................................QRPRDSEVEMMAQLLGIMVVASVCWLPLLVFIAQTVLRNPP.AMSPAGQLSRTTEKELLIYLRVATWNQILDPWVY PE2R4_HUMAN/34-329 GNLVAIVVLCKSRKEQK..ETTFYTLVCGLAVTDLLGTLLVSPVTIATYMKG........QWPGGQ...PLCEYSTFILLFFSLSGLSIICAMSVERYLAINHAYFYSHYVD..KRLAGLTLFAVYASNVLFCALPNMGLGSSRLQYP............DTWCFIDWTTNVTAHAAYSYMYAGFSSFLILATVLCNVLVCGALLRMHRQFMRRTSLGTEQHHAAAAASVASRGHPA.......................................................................................................ASPALPRLSDFRRRRSFRRIAGAEIQMVILLIATSLVVLICSIPLVVRVFVNQLYQPS.......LEREVSKNPDLQAIRIASVNPILDPWIY
As above you can perform alignment between the chosen pHMM and our first 30 up to 347 OR sequences. The MATLAB function hmmmerge allows an easier viewing.
for i=1:30 [Score(i), Seqs(i).Aligned] = hmmprofalign(hmm7tm, orseqs(i).Sequence); end hmmprofmerge(Seqs,Score)
It must be pointed out why it is better to use profile HMM instead of a pairwise alignment. Each time you align a sequence to an HMM, it is like aligning it to the hundreds of sequences that have been used to create the HMM. This gives you more certainty in the results of the alignment. As confirm, you can consider the following. The family to which we aligned the OR is the Rhodopsin family. Here you can try to perform an alignment with just one of the sequences used to develop the HMM. First, you must retrieve the sequences for the rhodopsin receptor and one of the odorant receptors.
rhod = getgenpept('NP_002368','sequenceonly',true); or = orseqs(300).Sequence;
Now you can perform a paiwise global alignment of the two sequences. The BLOSUM30 is used as scoring matrix. Here the penalties for opening and extending a gap in the alignment are set to 5.
[Score, Alignment] = nwalign(or, rhod, 'scoringmatrix','blosum30','gapopen',5,'extendgap',5)
Score = 54.8000 Alignment = MDVG-N-KS-TMSE--FVLLG--LS--NS-WELQMFFFMVFSLLYVATMVGNSLIVITVIVDPHLHSPMYFLLTNLSIIDMSLA--SFA-TPKMITDY-L-TGH-KTISFDGCLTQIFFLHLFTGTEIILLMAMSFDRYIAICKPLHYASVISPQVCVALVVA-SWIMGVMHSMSQVIFALTLPFCGPYEVDSFFCDLPVVFQLACVDTYVLGLFMISTSGIIALSCFIVLFNSYVIVLVTVKHHS-SRGSSKALSTCTAHFIVVFLFFG-PCIFIYM-W-PL-SSFLTDKILSV-FYTIFTPTLNPIIYTL-------RNQE-VKIAM-RKLKN----RF--LNFNKAM-PS-- || | | | :::| : :| | |: :::::: ::: |: |: | |: |:: :: : :| :::| |||:|:|| |: : : ::|| | :|| || : |: :|:: : || : ||:|:|::| :::: |: | |:: ||| | |::: : :: :: :::: | :| :: : : :| : |: :| |:: :::::: :: |::| ::| | :: : : ||| : : :: |::||:|: | ::|: : :: |:| : :|: | || : : ||:|| : | :| :|::: |::|: | | | :: :: MD-GSNVTSFVVEEPTNISTGRNASVGNAHRQIPIVHWVIMSISPVG-FVENG-ILLWFLCFRMRRNPFTVYITHLSIADISLLFCIFILSIDYALDYELSSGHYYTI-VT--LSVTFLFGYNTG--LYLLTAISVERCLSVLYPIWY-RCHRPKYQSALVCALLWALSCL--VT-TM-EYVM--C----IDR--EE-ESHSRNDCR-A-VI-IF-IAILSFLVFTPLM-LVSSTILV-VKIRKNTWASHSSKLYIVIMVT-IIIFLIFAMPMRLLYLLYYEYWSTFGNLHHISLLFSTINS-SANPFIYFFVGSSKKKRFKESLKVVLTRAFKDEMQPRRQKDNCNTVTVETVV
Now you must assess the significance of this alignment. Use randperm for this and compare the alignment and scores. The scores are fairly close. It is difficult to tell if the alignment is significant.
perm = randperm(length(or)); randor = or(perm); [Score, Alignment] = nwalign(randor, rhod,'scoringmatrix','blosum30','gapopen',5,'extendgap',5)
Score = 43.8000 Alignment = LLLASVTYGMNSAGTE-CTFVLMPVAN-HIQVLVKSDH-IKMSIEIVLKPCIKSVGYMPTCSDV--SPIFTFFADYVIWIAVVEALV-NLISYVGMKLPFFFLSN-VF-INIFSLNHMQHYIADLLLHTVMIHFTVFQVFIAFFKTTMRPSNPVL-TIKICGFGMMFYFHFNFTIVFSAISVLMTSTQASSSDSPSKISLLGSFICVYLSTMNFLPTFLVFDCDYAYQSMGCLRAVSLIVSPCETALSFIVVIRM-FSLSG-CKKLTEV-IWQPFILWLHI-FL-DLI-T---PLA-FML-TDL---Y--S--V-LTFGLNPDGLGYRMLRATL-THSIMSKV : ::|| : :: | | : |:| | |: : | : |||: | :: : : :| : :| ||::: :: || :: |: ::| : :| : : |: :: | ::|:: : |:: |:| | :| :| : : ::::: | | : :: :| :::: :: :| ::::::| ::::| | :: : || : || : |:|: || : :::: : ::| | |:: ::: ::::| : |:: : |: : | :| || :| ::| : |:: | : :: : | | || :: :| : |: :::: | :: : | MDGSNVTSFVVEEPTNISTGRNASVGNAHRQIPI-V-HWVIMSISPVGFVENGILLWF-LCFRMRRNP-FTVYITHLS-IADISLLFCIFILSIDYALDYELSSGHYYTIVTLSVTFLFGYNTGLYLLT-AI--SVERCLSVLYPIWYRCHRPKYQSALVC---ALLW-ALS-CLV-TTMEYVMCIDREEESHSRNDCRAVIIFIAI-LSFLVFTPLMLVSSTILVVKIRKNTWA-SHS-SKLYIVIMVTIIIFLIFAMPMRLLYLLYYEYWSTFGNLHHISLLFSTINSSANPFIYFFVGSSKKKRFKESLKVVLTRAF-KDEMQPRRQKDNCNTVTVETVV
On the contrary, if you align both sequences with the HMM, the significance of the alignment is more apparent.
[score_or, align_or] = hmmprofalign(hmm7tm,or) [score_rand, align_rand] = hmmprofalign(hmm7tm,randor)
score_or = 156.9790 align_or = GNSLIVITVIVDPHLHSPMYFLLTNLSIIDMSLASFATPKMITDYLTG-HKTISFDGCLTQIFFLHLFTGTEIILLMAMSFDRYIAICKPLHYASVIS-PQVCVALVVASWIMGVMHSMSQVIF-ALTLPFCGPYEVDSFFCDLPVVFQLACVDTYVLGLFMISTSGIIALSCFIVLFNSYVIVLVTVKHHSS--------RGSSKALSTCTAHFIVVFLFFGPC-IFIYMWPLSsFL--------------TDKILSVFYTIFTPTLNPIIY score_rand = -136.5222 align_rand = AVVEALVNLISYVGMK-LPFFFLSN---------VFINIFSLNHMQH-------YIADLLLHTVMIHFTVFQVF------------IAFFKTTMRPSN-PVLTIKICGFGmmfyfhFNFTIVFSAISVLMTSTQASSSDSPS--KISLLGSFICV----------YLSTMNFLPTFLVFDCDYAYQSMGCLRAVSLIVSPC----------ETALSFIVVIRMFSLSGCKKLTEVIWQPFIlWLHI-----FLDLITPLAFMLTDLYSVLTFGLNPdgLGY
Therefore, the alignment with pHMM has much more power than pairwise alignment since it includes the characteristics of all the sequences used to create the model.
Phylogenetic Tree
In the last part of this demo, you will create a phylogenetic tree from member of this protein family. The olfactory receptors are actually part of a much large protein family known as the G-Protein-Coupled Receptors. All of these proteins are 7-transmembrane, but they detect molecules other than odorants. There are 5 main groups of GPCRs: Adhesion, Secretin, Glutamate, Frizzled/TAS2, Rhodopsin (Fredriksson, et al.,2003). You will use a few of these groups to create the tree. First, sequences can be retrieved from the GenBank database using the getgenbank function.
data = {'Adhesion 1' 'NP_001775';
'Adhesion 2' 'NP_001965';
'Glutamate 1' 'NP_000830';
'Glutamate 2' 'NP_000836';
'Rhod-Alpha 1' 'NP_001051';
'Rhod-Alpha 2' 'NP_000946';
'Rhod-Delta 1' 'NP_002368';
'Rhod-Delta 2' 'NP_473372'};
for prot = 1:8
seqs(prot).Header = data{prot,1};
seqs(prot).Sequence = getgenpept(data{prot,2},'sequenceonly','true');
end
You can calculate the UPGMA distances using Jukes-Cantor correction, so you build the tree.
distances = seqpdist(seqs,'Method','Jukes-Cantor'); tree = seqlinkage(distances,'UPGMA',seqs)
Now plot the tree.
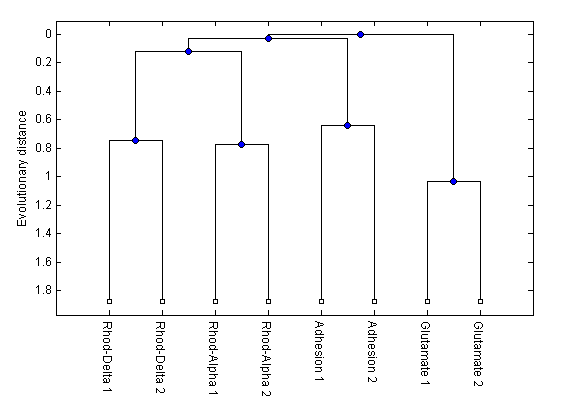
h = plot(tree,'orient','bottom'); ylabel('Evolutionary distance')
Adding two of the olfactory receptors sequences, you must recreate the tree.
data2 = {'Olfactory 1';'Olfactory 2'};
for prot = 1:2
seqs(prot+8).Header = data2{prot,1};
seqs(prot+8).Sequence = orseqs(prot).Sequence;
end
distances = seqpdist(seqs,'Method','Jukes-Cantor');
tree = seqlinkage(distances,'UPGMA',seqs)
h = plot(tree,'orient','bottom');
ylabel('Evolutionary distance')
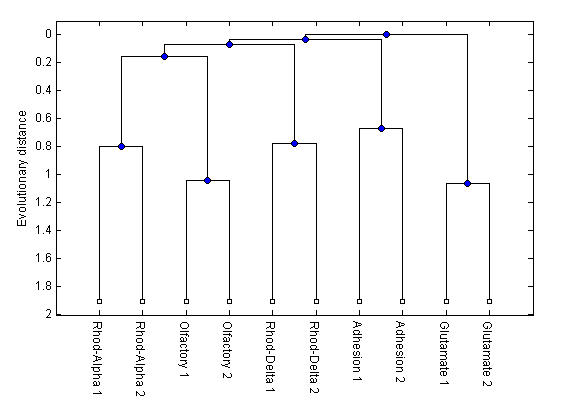
You can see that the members of the GPCR groups were grouped together and that the olfactory receptors fell within the Rhodopsin group. This matches what we previously knew from matching the ORs with the correct profile HMM. However, UPGMA grouped the ORs with the Alpha-Rhodopsins while the maximum parsimony method used by Fredriksson et al. group them with the Delta-Rhodopsins.
References
Axel, R. The Molecular Logic of Smell. 1995. Scientific American 273(4):154-159.
Buck, L. and R. Axel. 1991. A Novel Multigene Family May Encode Odorant Receptors: A Molecular Basis for Odor Recognition. Cell 65:175-187.
Eddy, S. Profile Hidden Markov Models. 1998. Bioinformatics. 14(9):755-763.
Fredriksson, R., M. Lagerstrom, L. Lundin and H. Schioth. 2003. The G-Protein-Coupled Receptors in the Human Genome Form Five Main Families. Phylogenetic Analysis, Paralogon Groups, and Fingerprints. Molecular Pharmacology 63:1256-1272.
Mombaerts, R. 2004. Genes and Ligands for Odorant, Vomeronasal, and Taste Receptors. Nature Reviews 5:263-278.
Zozulya, S., F. Echeverri and T. Nguyen. 2001. The human olfactory receptor repertoire. Genome Biology 2(6):research0018.1-0018.12.